‘The entire protein universe’: AI predicts shape of nearly every known protein
DeepMind’s AlphaFold tool has determined the structures of around 200 million proteins.
According to the “Guardian” report on July 28, Google’s artificial intelligence company DeepMind has further cracked almost all known protein structures. The database constructed by its AlphaFold algorithm now contains more than 200 million known protein structures. Drugs or new technologies to tackle global challenges such as famine or pollution paves the way.
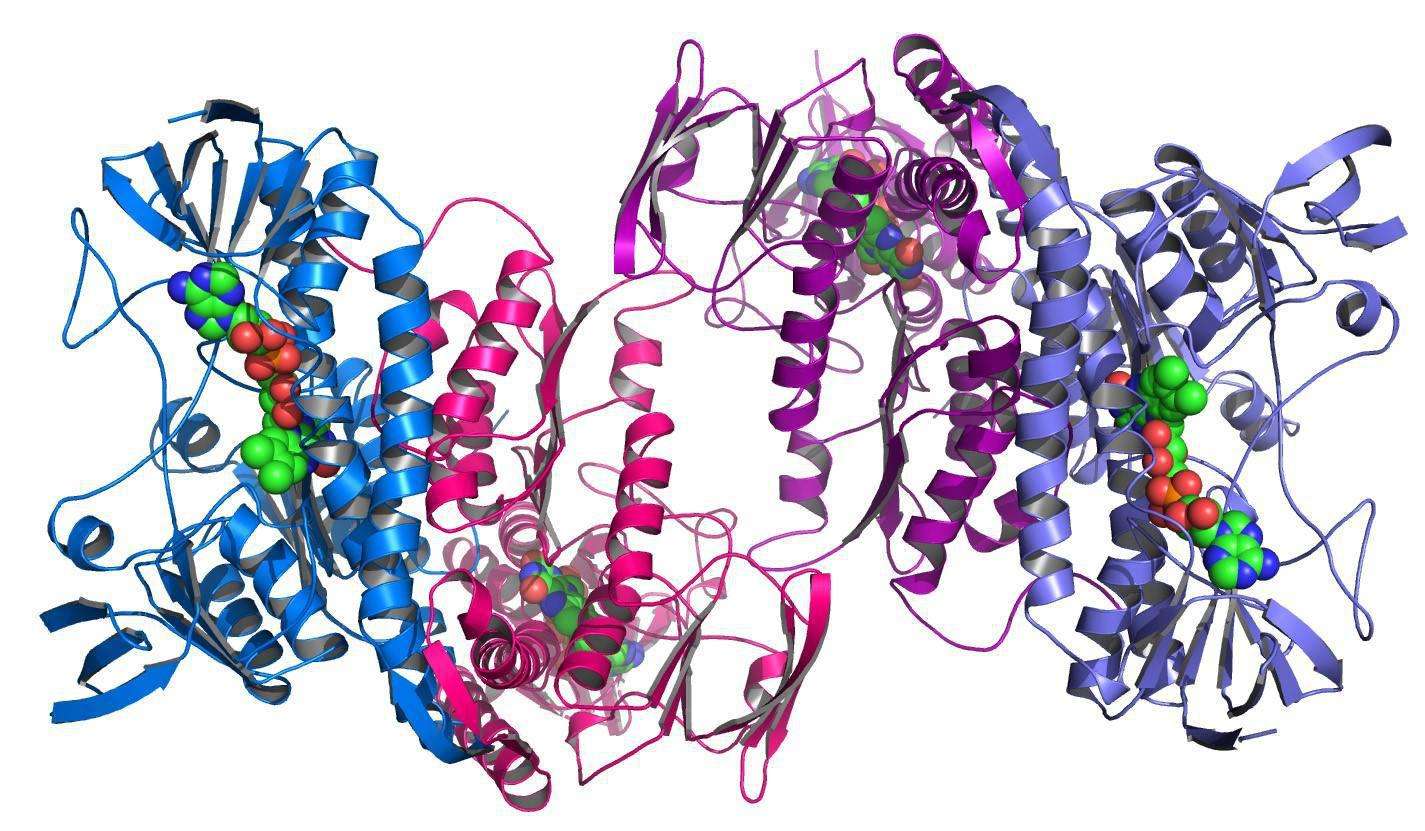
Proteins are the building blocks of life, and they consist of chains of amino acids that fold into different complex shapes. The function of a protein is often determined by its 3D structure. Once humans know how proteins fold, they can understand how they work and how to change their behavior. Although DNA provides instructions for making chains of amino acids, predicting how they interact to form 3D shapes is tricky, and until recently scientists have only deciphered a fraction of the 200 million proteins known to science The ability of the AlphaFold algorithm to decipher almost known protein structures is invaluable.
A year ago, DeepMind released AlphaFold2, which predicted 2/3 protein structures with atomic-level accuracy, and jointly released AlphaFold DB, an open and searchable protein structure database with EMBL-EBI, to share this technology with the world, This initial set of databases includes 98% of all human proteins.
Now, the database is expanding to more than 200 million structures, covering nearly every organism on Earth whose genomes have been sequenced, DeepMind said in a statement.
Demis Hassabis, founder and CEO of DeepMind, said, “Since artificial intelligence created this powerful new tool, users can now find the 3D structure of a protein almost as easily as a Google search for a keyword. This opens up enormous space for AlphaFold to have profound implications for important scientific issues such as sustainability, food security and neglected diseases. We are now at the beginning of a new era of digital biology.”
DeepMind has open sourced AlphaFold’s code and published two in-depth papers in the journal Nature with more than 4,000 citations. In addition, DeepMind collaborated with EMBL-EBI to design a tool to help biologists use AlphaFold and co-published AlphaFold DB.
Before releasing AlphaFold, DeepMind consulted with more than 30 biological research experts to share AlphaFold with the world in a way that maximized potential benefits and minimized potential risks.
“We believe that AlphaFold is by far the most important contribution of artificial intelligence to advancing science,” Pushmeet Kohli, head of AI science at DeepMind, said in a statement. .”
In fact, scientists have already used some of AlphaFold’s early predictions to help develop new drugs. In May, researchers led by Professor Matthew Higgins of the University of Oxford, UK, announced that they had used AlphaFold’s model to help determine the structure of a key malaria parasite protein, and to find out what could stop the parasite Locations where the propagated antibodies may bind.
Professor Higgins said: “Previously, we had been using a technique called protein crystallography to calculate the structure of this molecule, but because the molecule was very unstable and moved around, we couldn’t grasp its structure. When the AlphaFold model is combined with experimental evidence, suddenly everything makes sense. This discovery will be used to design and improve vaccines to induce the most effective antibodies to block the virus.”
In addition, scientists at the Centre for Enzyme Innovation at the University of Portsmouth, UK, are using AlphaFold’s model to identify enzymes in nature that can be tuned to digest and recycle plastics. Professor John McGeehan, who led the work, said: “We’ve spent quite some time navigating through this vast database of structures and have discovered a range of new, never-before-seen plastics that can actually break down plastic. 3D shape. It was a huge success. AlphaFold’s model could speed up our research and help us put these precious resources into important research.”
Professor Janet Thornton, group leader and senior scientist at the European Institute of Bioinformatics at the European Molecular Biology Laboratory, also pointed out, “AlphaFold protein structure prediction has been used in many ways. I expect that this latest Advances will lead to a wealth of new and amazing scientific discoveries in the months and years to come, all thanks to the fact that the data is public and available to all.”
To date, more than 500,000 researchers from 190 countries have accessed AlphaFold DB, viewing more than 2 million structures. Some freely available protein structures have also been integrated into other public datasets, such as Ensembl, UniProt, and OpenTargets, accessed by millions of users.